Advanced Query
Simple query does the path finding for you. If you want more control, use the Advanced query.
To make an advanced query, you will need to know,
- Data that you already have (e.g. T-cell receptor sequence)
- Data that you want to find (e.g. specific peptide-MHC complex that the T-cell receptor binds to)
- The path to get from your data to the data you want to know
Depending the path that you specify, the data returned would mean different things scientifically. Refer to the query path guide
Basics of query pathing
For example, if you want to find the MHCs which bind to the Tcr.
The schema of the modules is as follows (more details here):
Therefore, the path to find the peptide-MHC complexes that bind to your TCRs would be,
Preparing your data
To make your query, you first have to prepare your CSV file containing your Tcr data.
For example, you have some CSV data of TCRs,
| VBeta.gene | VAlpha.gene | JAlpha.gene | Cdr3Alpha.id |
|---|---|---|---|
| TRBV14*01 | TRAV26-1*01 | TRAJ37*01 | CIVVRSSNTGKLIF |
| TRBV13-2*01 | TRAV6D-3*01 | TRAJ13*01 | CAANSGTYQRF |
| TRBV6-3*01 | TRAV29/DV5*01 | TRAJ52*01 | CAASVYAGGTSYGKLTF |
Building your query
On the app, change the mode from 'Simple Query' to 'Advanced Query':
Then, upload your TCR data, as per normal.
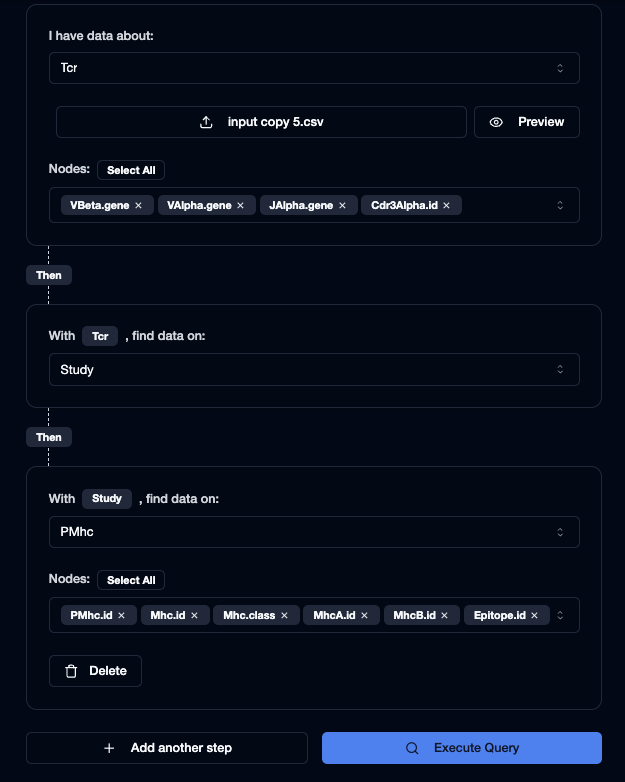
Next because we have 3 steps in our path, we need to add the 2nd and 3rd step via the "Add another step" button.
Then, set the 2nd step as Study, and set the 3rd step as PMhc. On the 3rd step, under "Nodes" press "Select All" to find out all data about the peptide-MHC complex.
This is what your query should look like:
Lastly, press execute query to view your results.